Data
Processing
The Xplain software module
designed for processing and interpretation the data obtained form microarrays
images. The program works with files created by the Xplore image-processing
module. Selecting the FileàOpen menu
item opens the standard file open dialog.
It is necessary to select the
desired file with integral intensity data created by the Xplore module and
press the <Open> button. The process of opening the file may take a few
seconds because all the necessary calculations done by the program at this
stage. After the file has opened the main window of the Xplain looks in the
following way:
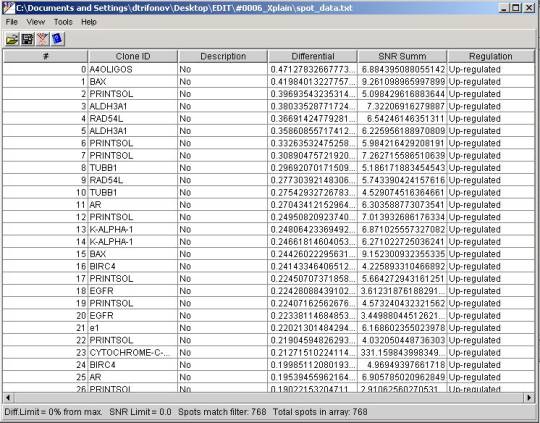
In the table that appeared as a
result of opening the file each of the rows corresponds to the spot at the
microarray. Meanings of the columns are following:
- # - serial number of a row in the table
- Clone ID - clone name the same
that has been registered in the source microplate
- Description - non-mandatory
description field transferred form the microplate registered
- Differential - value of the
differential expression of the clone in the experiment
- SNR Summ - sum of SIGNAL to
BACKGROUND ratios in the first and second scanning channels
- Regulation - positive
differential represents an Up-regulated gene, negative is Down-regulated
Sorting.
By default the rows sorted by the
differential descending. To sort them in the alphabetical order select the
ToolsàFilter
menu item and in the opened filter dialog
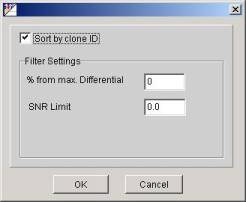
check the "Sort by clone ID" checkbox.
Calculations.
There are a number of parameters
taken from the integral intensity file used for calculation of the differential
expression of clones.
S1 - integral intensity of pixels
from the SIGNAL area in the first scanning channel
S2 - integral intensity of pixels
from the SIGNAL area in the second scanning channel
B1 - integral intensity of pixels
from the BACKGROUND area in the first scanning channel
B2 - integral intensity of pixels
from the BACKGROUND area in the second scanning channel
As - number of pixels in the
SIGNAL area
Ab - number of pixels in the
BACKGROUND area
Data Normalization.
Due to the difference in
sensitivity of the scanners' channels and difference in the probes labeling a
constant difference in the fluorescence level of the microarray surface in
different channels may occur. This difference can be partially compensated by
the normalization procedure:
The program calculates average
signal of all spots on the microarrays in the firs and in the second channels:
Savg1 è Savg2.
The normalization coefficient is
taken as a ratio of these values:
Knorm = Savg1/Savg2
Intensity of each spot in the
second channel multiplies by this coefficient:
S1norm = S1
S2norm = S2 x Knorm
Differential Expression
Calculation.
Differential = log( S1norm /
S2norm )
Signal to Background Ratio
Calculation.
The signal and the background
areas may contain different amount of pixels. Due to this it is necessary to
deal with the values of signal and background densities instead of dealing with
direct values:
Ds1 = S1/As - signal density in
the first channel
Ds2 = S2/As - signal density in
the second channel
Db1 = B1/Ab - background density
in the first channel
Db2 = B2/Ab - background density
in the second channel
SNR summ = Ds1/Db1 + Ds2/Db2
All above calculations take place
for each spot on the microarray.
The Xplain presents data that
helps making conclusions regarding quality of the experiment.
The ToolsàQC menu
item opens the QC dialog window.
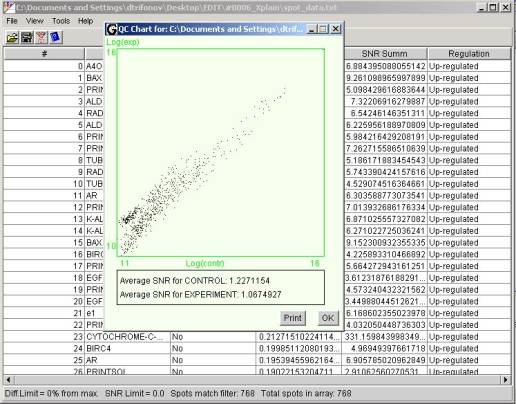
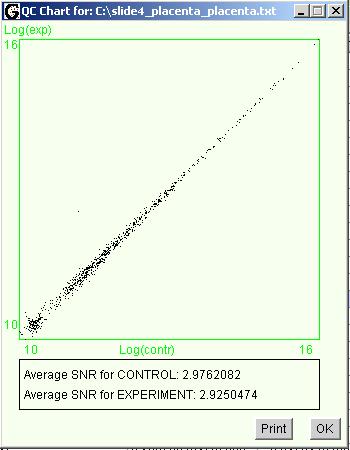
In the upper part of the window
resides the scatter plot. Values along the X and Y axes are logarithms of
normalized intensities in the first and in the second scanning channels
correspondingly. Each spot on the microarray represented by a dot on the
scatter plot.
It can be
the following interpretation to the scatter plot. Most of the spots at a
reasonably large microarray have close to zero differentials. Therefore ideally
most of the scatter plot dots suppose to be concentrated along the diagonal
line coming from the left lower corner of the chart area. All deviations of
spots from the diagonal can be caused either by the differential expression or
by an error in the experiment. If most of the spots scattered away from the
diagonal that picture can be interpreted, as multiple errors exist in the
experiment. In the particular example shown at the picture above scattering of
the dots can be explained by a rather low signal to background ration of the
whole experiment. Average values of the signal to background ratios for both
channels shown under the chart area in the QC window. The fact that apparently
most of the spots reside over the diagonal indicates that the numerical
normalization of the data was not enough to correct the difference in the
channels completely. It would be expedient to re-scan the microarray and do
channels leveling based on the background integral intensity. This information
can be obtained using the Xplore statistical analysis tools. Noticeable
aggregation of spots in the left lower corner slightly apart from the rest of
the spots can be caused buy a runs or drops of washing liquid dried on the
microarray surface affecting it's uniformity.
For comparison there is a chart of
a microarray data obtained from the high quality experiment and processed in a
proper way. In spite of a very low signal to background ratio, around 3, the
data has small dispersion and therefore high reliability and repeatability.
One spot placed significantly away
from the rest of the spots conglomerate corresponds to a prelabeled control
clone that supposed to have a large differential.
Not all spots may be of interest
to an experimenter. It makes sense to
exclude from consideration all spots with too low signal to background ratio
because they provide unreliable data. Signal to background credibility
threshold differs from one experiment to another. It can be determined by
comparing data from spots duplicates. And that is the reason to duplicate spots
on the array whenever it's possible in order to have the comparison basis. It
is known from the experience that in the most of experiments spots with the
signal to background ratio over 5-6 give credible information.
In order to set the S/B threshold
and let the program filter out all spots below that level select the ToolsàFilter
menu item and in the opened filter dialog enter the desired value in the
"SNR Limit" field.
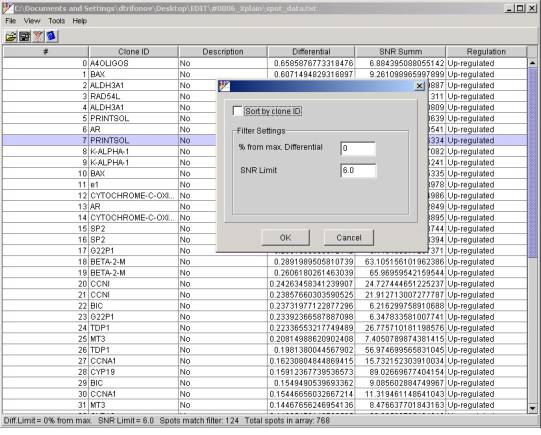
After that has done the table
reflect only rows that match the filter criterion. The status bar at the Xplain
main window shows how many spots have been selected. In this example for the
S/N threshold equal to 6:
Spots match filter: 124-selected
124 spots
from
Total spots in array: 768.
Differential Expression Level
Selection.
The second parameter of the filter
defines the differential expression threshold in the percentage from the
maximal differential expression found at a particular microarray. The 0 value
of this parameter allows all spots to be shown, the 100 value results in
selection of only one spot the one that has the highest differential expression
level. The two filter parameters liked
with the logical "AND" function therefore selected data must comply
with both criteria of the filter.
In the example following filter
settings:
% from max. Differential = 90
SNR limit = 6.0
will drop off all rows. It means
that spots with the highest differential expression do not mach the S/N
criterion and provide unreliable data.
Data Exporting.
The table data can be saved in a
.TXT file for further processing by another applications. Use the FileàSave As
menu item in order to do this.
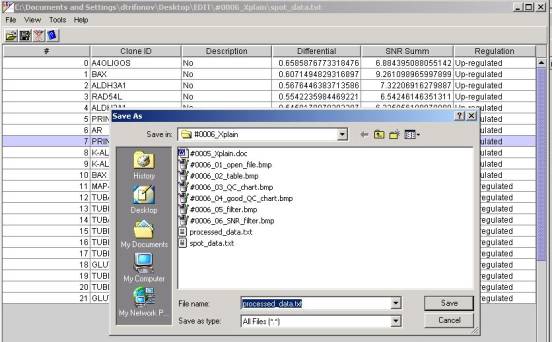
Only the rows that match filer
criteria will be placed in the file. File structure is identical to the
structure of the table. Columns separated by the " , " (comma)
symbol.