Microarray
Image Processing.
Image processing is done with the
Xplore program. Input for this program are arrays images produced by scanner
and annotation file produced by the spotter. The Xplore output is a file
containing values of integral fluorescence of the surface of the microarrays
carrying the DNA spots. Besides this the Xplore offers a number of tools for
visual and statistical analysis of the substrate surface.
Image Loading.
Selection of the FileàOpen menu
item opens the standard file open dialogue. The dialogue shows files with TIFF
extension because only that standard supported by the Xplore. Select desired
file and press the <Open> button.
After the file is opened the
Xplore main window looks in the following way:
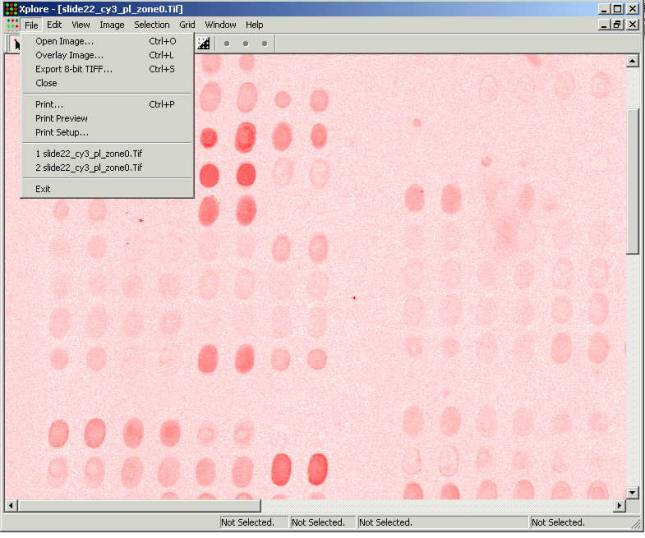
Image of the first opened image is
being drawn by the red color. (Please note that a microarray of rater low
quality has been selected for this manual in order to demonstrate all
advantages of the Xplore. The microarray selected contains printing, processing
and scanning defects yet it is still possible to extract meaningful data out of
this experiment.)
Composite Image.
When the first image was opened a
few additional items were added to the Fileà… menu.
- Overlay Image - allows overlaying another images made in the
other scanner channels over the one that has already opened.
- Export 8-bit TIFF - allows to
save the image in the 8 bit TIFF format that is compatible with the majority of
commercial graphical software. This file is intended for the purposes of
presentations and publication of experimental materials and does not contain
any quantitative information.
Selecting the FileàOverlay
Image opens the same file open dialogue. It is important to open only images of
the same microarray obtained in the other scanning channels for overlaying.
Otherwise the program reports an error and prevents the incorrect image from
being overlaid over the first one. If the image overlay operation went
successfully the Xplore window starts looking in the following way:
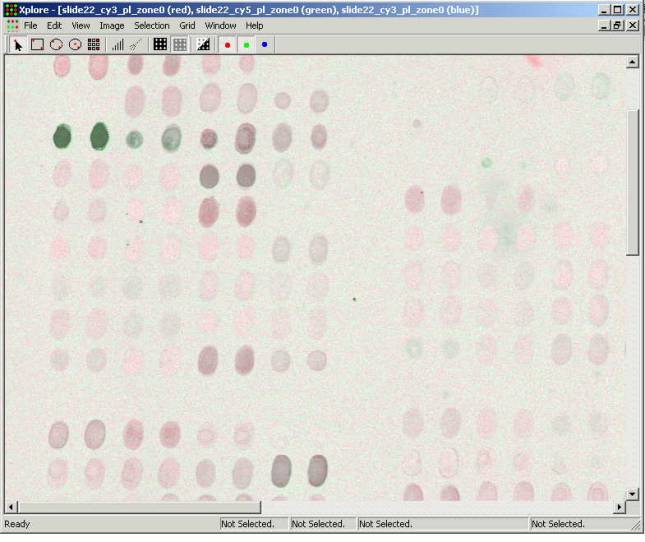
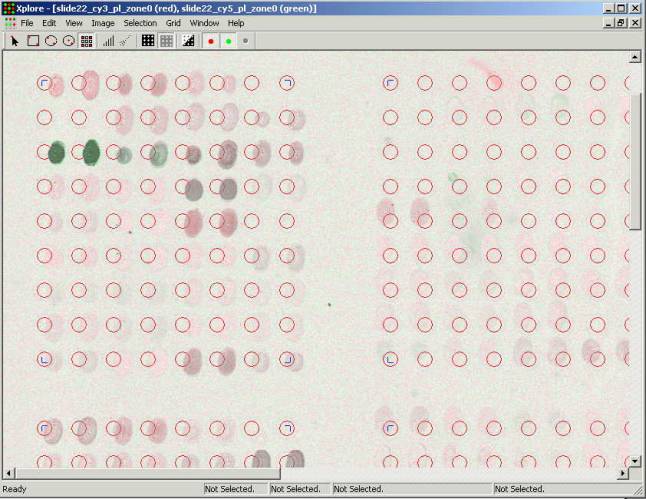
The second opened image is being
drawn with the green color. The Xplore allows three images to be overlaid. The
third overlaid image will be in the blue color. (All examples in this manual
were prepared using two images obtained by scanning a microarray in the
channels that correspond to Cy3 and Ñy5 fluorescent dyes). Pressing buttons with colored dot at the
toolbar turns on and off corresponding images. When all buttons are pressed the
images are overlaid forming the composite image. When images overlay the colors
mix proportionally to the intensity of the surface elements (pixels). If
intensity is equal in both channels this area is being shown gray. Intensity
level of the gray color corresponds to the level of intensity of the
surface. Prevailing of the red or green
color indicates the difference of fluorescence intensity in the channels. Color intensity is proportional to the
fluorescence intensity in the corresponding channel. It is correct to assume
that colored spots carry clones that expressed differently in this experiment
and the gray ones have no differential expression.
Applying the Grid.
The grid defines zones of data
extraction. In order to perform proper data extraction it is necessary to place
the grid cells over the corresponding spots on the images.
Go to GridàDefine
Grid menu item to open the Grid Parameters dialogue.
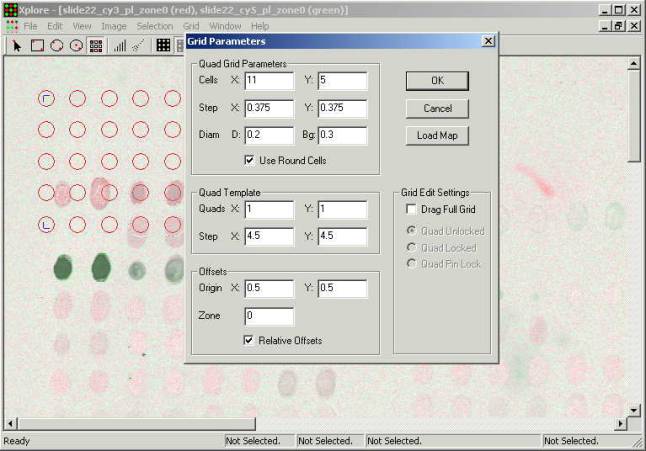
It is possible to define grid
manually or load a file with microarray layout generated by the spotter. A grid
done manually does not contain annotation of the clones, while the grid that is
taken form the generated file contains both annotation of clones on the chip
and their positions.
In order to load a file with grid
parameters generated by the spotter press the <Load Map> button in the
Grid Parameters dialogue window. At the standard file open dialog select the
appropriate file. The Xplore main window after the grid has been applied shown
below:
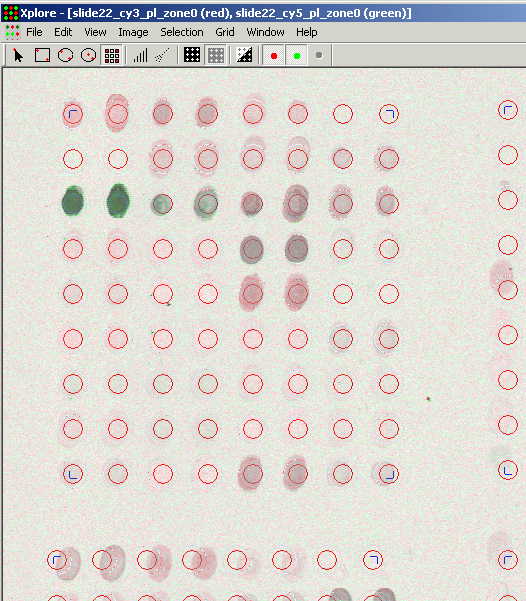
Grid cells approximately fit
positions of the spots. The objective of the further operations with the grid
is to place the cells exactly over the spots. Average time requires to position
the grid over a 1000 clones array is about 10 minutes, however for larger array
of a number thousands spots this may take a significant time. In order to make interruptions in this
continuous process saving intermediate parts of the work it is possible to save
edited grid in a file. To do this select the GridàSave Grid menu item. The
grid will be saved in a file with .grd extension. In order to get back to the
same image open the saved grid file not the one that was originally produced by
the spotter.
Manual Grid Adjustments.
The Xplore does not allow moving
each cell individually. That would have been too laborious indeed. There are
alternative means built in the program that allow adjusting the grid fast and
accurately.
In order to move all cells of the
grid simultaneously move the pointer to a blue angle in one of the corners of
the quads of spots in the array. Holding the <Shift> button at the
keyboard and the left button of the mouse move the entire quad of the grid cells
all the way to combine the left upper cell of the grid with the left upper spot
in the array.
After that select the GridàDefine
Grid menu item and in the opened window "Grid Parameters" press the
<OK> button doing no other actions. This will result in shifting of all
of the grid cells at the same distance and in the same direction as the first
quad did. If there are no printing defects at the chip this may be the last
step required to combine the grid cells with the corresponding spots. However
if the microarray has been printed with some quads off the rectangular
formation it is necessary to process each of the quads individually.
It is possible to distort the
rectangular form of the grid if it's necessary to reflect some distortions of
the pattern the spots have printed. Left click on a one of the blue angles in
the quads corners and drag it without holding the <Shift> key. The grid
cells will move as if they attached to a rubber thread.

That is the way of dealing with
distortions inside a quad. For the vast majority of images this procedure is
enough to place all cells accurately over the spots and proceed to the data
extraction.
Automated Grid Adjustment.
In case of major printing defects
it is possible to apply one of the automated algorithms that designed to locate
spots and move each of the cells individually. In order to do this it is
necessary to perform all above actions and place the grid manually with the
maximal possible accuracy. After that select one of the menu items: GridàAdjust
Grid 2DW or GridàAdjust
Grid 1DW or GridàAdjust
Grid CM. The program place grid cells in the areas of the maximum intensity.
Use the algorithm that brings the best results for a particular image/
Please note that due to
independent movements of the grid cells some of the cells can be placed
significantly asymmetrically.
Data Extraction Zones.
The process of data extraction
deals with integrating of intensity all pixels circumscribed by the grid cells
borders for each of the scanning channels apart and saving the integral
intensity values in a file that can be used by the data analysis software.
There are zones of integration of the intensity take place
during the data extraction - the SIGNAL zone and the BACKGROUND zone. Knowledge
of the background level around spots gives indirect information regarding
quality of the experiment. Each of the cells drawn as two concentric circles:
red and blue. These circles limit the signal and the noise zones
correspondingly for a particular cell. The ring between the circles is a border
zone. No data extracted out of it.

The default signal zone diameter
is equal to the diameter of the printing needles used for this microarray. The
default background zone diameter is 1.5 times bigger. It is possible to change
those parameters manually. To do so open the Grid Parameters dialogue following
the GridàDefine
Grid menu item.
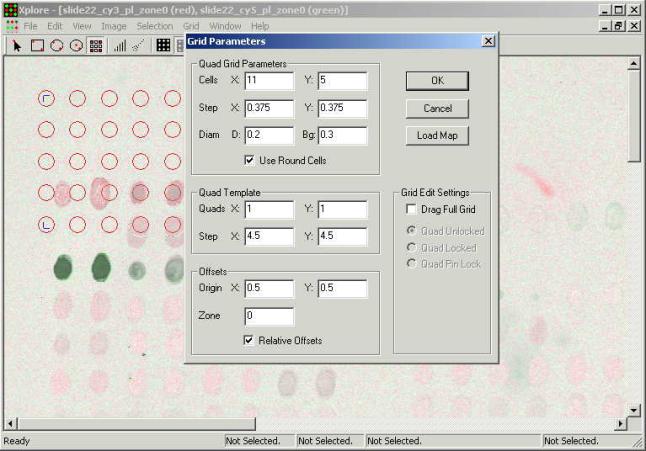
Data fields next to the
"Diam." label define diameters of the zones "D:" of the
signal and "Bg:" of the background. The rest of the fields in this
dialogue mean following:
Quad Grid Parameters
- Cells X: Y: number of grid cells
along the X and Y axes.
- Step X: Y: distance between
cells along the X and Y axes.
- Use Round Cells option of using
round cells vs. squared ones. (in case of the squared cells diameter means side
of the square)
Quad Template
- Quads X: Y: number of spot quads
along the X and Y axes.
- Step X: Y: distance between
quads (typically equal to the distance between the needles)
Offsets:
- Origin X: Y: position of the low
left spot on the chip relative to the low left corner of the substrate.
- Zone - printing zone number in
case of multiple zones.
- Relative Offsets - adds zone
offset specified in slide description file produced by printer program.
To extract the data select the
GridàExtract
Grid Data menu item. In the opened standard file saving dialog type the name of
the integral intensity data file and press the <Save> button. This file
has can be used by the Xplain data processing program or by a third party
software such as Excel or Mathlab.
Clones Search.
Combination of the <Ctrl>
and <F> at the keyboard opens the search window.
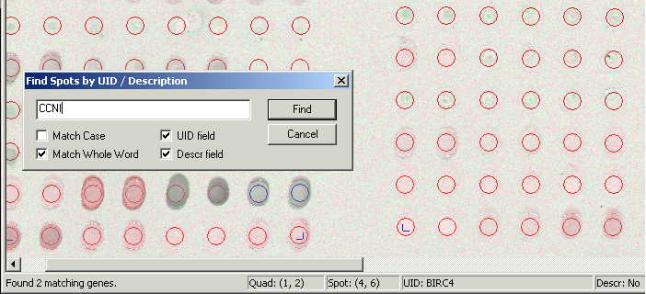
Enter the name of the clone or a
part of the name in the search field. If the exact name is known mark the
"Match Whole Word" checkbox, otherwise mark the "Match
Case" checkbox. The other two checkboxes in the dialog window define the
fields the desired annotation will be searched in. Search results shown in the
left side of the status bar indicating the number of occurrences found in the
search. All the corresponding cells are being highlighted in the same time with
the blue circle. (The circle line is single width compare to manual
selection). In the example shown at the
picture above two clones were found which is witnessed by the "Found 2
matching genes" message. The spots have found located in the middle of the
second row from the bottom.
Location of Low Intensity Spots.
It is typical to deal with low
intensity images in the most of the microarrays experiments. Signal to
Background (S/B) ratio of low intensity spots is less than 10. However even
spots with signal to background ration around 5 may bring reliable information.
Spots with such a S/B ratio can be barely recognized on the image. It might be
difficult to locate them and place the grid correctly. The Xplore provides
certain tools to locate the low intensity spots.
Image Inversion.
The ![]() button inverses the image.
button inverses the image.
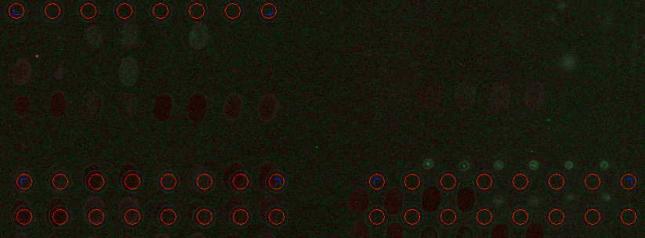
In some instances low intensity
spots appear more contrast on dark background. Inversion changes the picture
only and does not affect the data.
Background Subtraction
One of the ![]() buttons starts the selection mode. The shape
of the zone is being selected corresponds to the picture at a particular
button. It is necessary to select a background area between the spots and go to
the ImageàSubtract
Background menu item. From intensity of each pixel the program will subtract
average intensity of the selected zone. The image will become lighter (darker
in the inverse mode) and this may result in increase in contrast.
buttons starts the selection mode. The shape
of the zone is being selected corresponds to the picture at a particular
button. It is necessary to select a background area between the spots and go to
the ImageàSubtract
Background menu item. From intensity of each pixel the program will subtract
average intensity of the selected zone. The image will become lighter (darker
in the inverse mode) and this may result in increase in contrast.
Statistical Analysis.
The "Xplore" offers
means for basic statistical analysis of the selected zone. Pressing the ![]() button (or selecting the SelectionàShow
Selection Statistics menu item) opens the Selected Data Statistics dialog:
button (or selecting the SelectionàShow
Selection Statistics menu item) opens the Selected Data Statistics dialog:
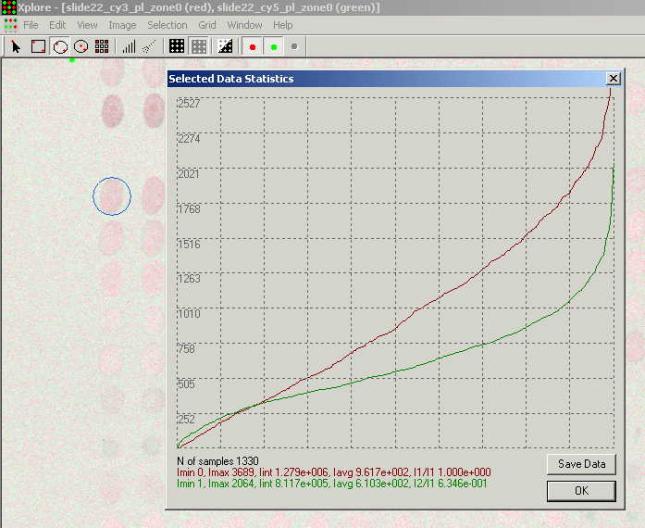
In the upper part of the opened
window shown the histogram of intensity of the pixels from the selected zone
for each of the scanning channels. The X axis shows number of pixels of a
particular intensity. The Y axis shows pixels intensity in the dynamic range of
the current selection zone. The red line represents data for the first opened
channel the green one for the second channel.
In the bottom part of the window
basic statistics is shown:
N of samples: 1330 - means that
the selected area contains 1330 pixels.
I min 0 - minimal intensity pixel
has intensity equal to 0 (1 for the green channel)
Imax. - maximal intensity pixel
Iint. - integral intensity for all
pixels in the selected area
Iavg. - average intensity for all
pixels in the selected area
I1/I1 ( I1/I2 )- ratio of integral
intensities of the pixels from two channels. Meaningless for one opened channel
- equal 1. If two channels are open - it is I1/I2; equal to 0.6346 in this
example.
Histogram data can be saved in a
txt file by pressing the <Save Data> button.
Equalizing
Sensitivity of Scanner Channels.
Due to the non-uniform labeling of
hybridization probes fluorescent levels of Cy3 and Cy5 dies may differ
significantly. It is important to adjust sensitivity of scanner channels to
obtain images with approximately equal average values of the intensity. We
recommend using the dialog described in the previous paragraph to obtain data
of the average intensity (Iavg.) for both channels. Select a large area of the
image that covers both background zone and the zone where spots reside. If
average intensity values of two channels of the area selected differ more than
10-15% from each other try to decrease sensitivity of the channel with the
higher intensity. Rescan the microarray with the new sensitivity settings.
Repeat measurements and rescanning till the both channels of the scanner
produce images with equal Iavg. values.
Scatter plot.
Pressing the ![]() button opens the Selected Data Pairs dialog.
button opens the Selected Data Pairs dialog.
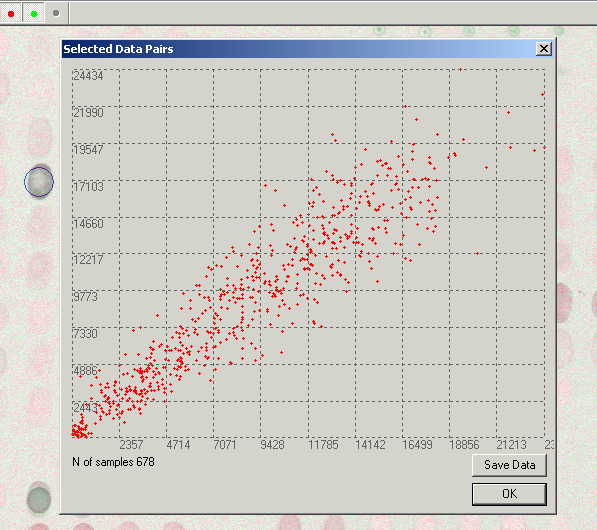
The X and Y axes show value of
intensity in the first and the second channels correspondingly. Spots at the
chart indicate intensities of pixels from the selected area.