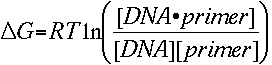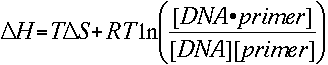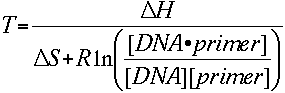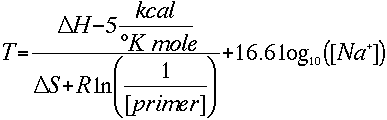Oligonucleotide Properties Calculator
To use this calculator, you must be
using Netscape 3.0 or later
or Internet Explorer version 3.0 or later, or
another Javascript-capable browser
Self-Complementarity requires a 4.x
browser. IE 5.0 is also supported.
This page was written in
Javascript.
Extensively rewritten from 12/15/2000-12/19/2000 to isolate
javascript Oligo object behaviors for teaching purposes.
This page may be
freely distributed for any educational or non-commercial use.
Copyright
Northwestern University, 1997-2002.
About the Calculations
Thermodynamic Calculations
The nearest neighbor and thermodynamic
calculations are done essentially as described by Breslauer et al.,
Proc. Nat. Acad. Sci. 83, 3746-50, 1986 (Abstract)
but using the values published by Sugimoto et al., Nucl. Acids
Res. 24, 4501-4505, 1996 (Abstract).
This program assumes that the sequences are not symmetric and contain at least
one G or C. The minimum length for the query sequence is 8.
The melting temperature calculations are based on the simple thermodynamic
relationship between entropy, enthalpy, free energy and temperature, where
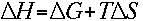 The change in entropy (order or a measure of the randomness of the
oligonucleotide) and enthalpy (heat released or absorbed by the oligonucleotide)
are directly calculated by summing the values for nucleotide pairs obtained by
Breslauer et al., Proc. Nat. Acad. Sci. 83, 3746-50, 1986.
The relationship between the free energy and the concentration of reactants and
products at equilibrium is given by
The change in entropy (order or a measure of the randomness of the
oligonucleotide) and enthalpy (heat released or absorbed by the oligonucleotide)
are directly calculated by summing the values for nucleotide pairs obtained by
Breslauer et al., Proc. Nat. Acad. Sci. 83, 3746-50, 1986.
The relationship between the free energy and the concentration of reactants and
products at equilibrium is given by
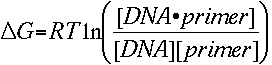 Substituting the two equations gives us
Substituting the two equations gives us
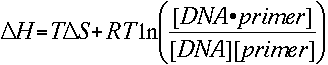 and solving for temperature T gives
and solving for temperature T gives
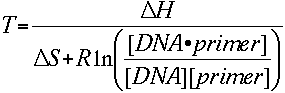 We can assume that the concentration of DNA and the concentration of
the DNA-primer complex are equal, so this simplifies the equation considerably.
It has been determined empirically that there is a 5 (3.4 by Sugimoto et al.)
kcal free energy change during the transition from single stranded to B-form
DNA. This is presumably a helix initiation energy. Finally, adding an adjustment
for salt gives the equation that the Oligo Calculator uses:
We can assume that the concentration of DNA and the concentration of
the DNA-primer complex are equal, so this simplifies the equation considerably.
It has been determined empirically that there is a 5 (3.4 by Sugimoto et al.)
kcal free energy change during the transition from single stranded to B-form
DNA. This is presumably a helix initiation energy. Finally, adding an adjustment
for salt gives the equation that the Oligo Calculator uses:
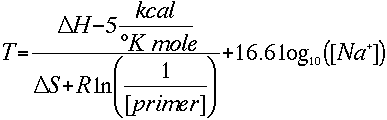 No adjustment constant for salt concentration is needed, since the
various parameters were determined at 1 Molar NaCl, and the log of 1 is zero.
No adjustment constant for salt concentration is needed, since the
various parameters were determined at 1 Molar NaCl, and the log of 1 is zero.
ASSUMPTIONS:
The thermodynamic calculations assume that the
annealing occurs at pH 7.0. The melting temperature (Tm) calculations
assume the sequences are not symmetric and contain at least one G or C.
The oligonucleotide sequence should be at least 8 bases long to give
reasonable Tms.
|
Basic Melting Temperature (Tm) Calculations
The two standard
approximation calculations are used. For sequences less than 14 nucleotides the
formula is
Tm= (wA+xT) * 2 + (yG+zC) * 4
where w,x,y,z are the number of the bases A,T,G,C in the sequence,
respectively.
For sequences longer than 13 nucleotides, the equation
used is
Tm= 64.9 +41*(yG+zC-16.4)/(wA+xT+yG+zC)
ASSUMPTIONS:
Both equations assume that the annealing
occurs under the standard conditions of 50 nM primer, 50 mM
Na+, and pH 7.0.
|
Salt Adjusted Melting Temperature (Tm) Calculations
A variation on two
standard approximation calculations are used. For sequences less than 14
nucleotides the same formula as the basic calculation is use, with a salt
concentration adjustment
Tm= (wA+xT)*2 + (yG+zC)*4 - 16.6*log10(0.050) +
16.6*log10([Na+])
where w,x,y,z are the number of the bases A,T,G,C in the sequence,
respectively.
The term
16.6*log10([Na+]) adjusts the Tm for
changes in the salt concentration, and the term
log10(0.050) adjusts for the salt adjustment at 50
mM Na+. Other monovalent and divalent salts will have an
effect on the Tm of the oligonucleotide, but sodium ions are much more effective
at forming salt bridges between DNA strands and therefore have the greatest
effect in stabilizing double-stranded DNA.
For sequences longer than 13 nucleotides, the equation used is
Tm= 100.5 + (41 * (yG+zC)/(wA+xT+yG+zC)) - (820/(wA+xT+yG+zC)) +
16.6*log10([Na+])
Symbols and salt
adjustment term as above, with the term (41 * (yG + zC-16.4)/(wA + xT +
yG + zC)) adjusting for G/C content and the term (820/(wA + xT
+ yG + zC)) adjusting for the length of the sequence.
ASSUMPTIONS:
Both equations assume that the annealing
occurs under the standard conditions of 50 nM primer and pH
7.0.
|
OD Calculations
Molar Absorptivity values in 1/(Moles cm)
| Residue |
Moles-1 cm-1 |
Molecular Weight
(after protecting groups are removed) |
| Adenine (dAMP, Na salt) |
15200 |
313.21 |
| Guanine (dGMP, Na salt) |
12010 |
329.21 |
| Cytosine (dCMP, Na salt) |
7050 |
289.18 |
| Thymidine (dTMP, Na salt) |
8400 |
304.2 |
| 6'
FAM |
20960 |
537.46 |
| TET |
16255 |
675.24 |
| HEX |
31580 |
744.13 |
| TAMRA |
31980 |
Assume 1 OD of a standard 1ml solution, measured in a cuvette with a 1 cm
pathlength.
| Chemical name: |
6-carboxyfluorescein |
| Absorption wavelength maximum: |
495 nm |
| Emission wavelength maximum: |
521 nm |
| Molar Absorptivity at 260nm: |
20960 Moles-1 cm-1 |
| Chemical name: |
4, 7, 2', 7'-Tetrachloro-6-carboxyfluorescein |
| Absorption wavelength maximum: |
519 nm |
| Emission wavelength maximum: |
539 nm |
| Molar Absorptivity at 260nm: |
16255 Moles-1 cm-1 |
| Chemical name: |
4, 7, 2', 4', 5', 7'-Hexachloro-6-carboxyfluorescein |
| Absorption wavelength maximum: |
537 nm |
| Emission wavelength maximum: |
556 nm |
| Molar Absorptivity at 260nm: |
31580 Moles-1 cm-1 |
TAMRA:
| Chemical name: |
N, N, N', N'-tetramethyl-6-carboxyrhodamine |
| Absorption wavelength maximum: |
555 nm |
| Emission wavelength maximum: |
580 nm |
| Molar Absorptivity at 260nm: |
31980 Moles-1 cm-1 |
Nucleotide base codes (IUPAC)
| Symbol: nucleotide(s) |
| A |
adenine |
| C |
cytosine |
| G |
guanine |
| T |
thymine in DNA;
uracil in RNA |
| N |
A or C or G or T | |
| M |
A or C |
| R |
A or G |
| W |
A or T |
| S |
C or G |
| Y |
C or T | |
| K |
G or t |
| V |
A or C or G; not T |
| H |
A or C or T; not G |
| D |
A or G or T; not C |
| B |
C or G or T; not A | |
Most recent version is available at URL: http://www.basic.northwestern.edu/biotools/oligocalc.html
The current version is the result of efforts by the following people:
Qing Cao, M.S. e-mail
Research
Computing
Northwestern University Medical School
Chicago, IL 60611
Warren A. Kibbe, Ph.D. e-mail and PH entry.
Research Computing
Northwestern University
Medical School
Chicago, IL 60611
Original code by Eugen
Buehler
Research Support Facilities
Department of Molecular Genetics
and Biochemistry
University of Pittsburgh School of Medicine
Monomer structures and molecular weights provided by Bob Somers, Ph.D.
Sr. Applications
Chemist
Glen Research Corporation
22825 Davis Drive
Sterling, VA
20164
http://www.glenres.com/
Uppercase/lowercase strand complementation problem described by Alexey Merz
alexey@dartmouth.edu