Covalent binding is established using the condensing reagent: 1-Ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) as the coupling reagent (
1. Introduction
Covalent binding is established using the condensing reagent: 1-Ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) as the coupling reagent ( ![]() ). The covalent binding of the phosphorylated solid phase oligonucleotide to the NucleoLink Strips is a very important step in the DIAPOPS procedure. The DIAPOPS binding procedure (
). The covalent binding of the phosphorylated solid phase oligonucleotide to the NucleoLink Strips is a very important step in the DIAPOPS procedure. The DIAPOPS binding procedure ( ![]() ) has been designed to bind the highest number of oligonucleotides as rapidly as possible on the basis of results from numerous optimization experiments, taking both price and simplicity of the method into consideration. Results of these optimization experiments are summarized in the following text. The term "solid phase oligonucleotide" is used in this text instead of "solid phase primer" since the solid phase bound oligonucleotide does not function as a primer unless the DIAPOPS procedure is carried out.
) has been designed to bind the highest number of oligonucleotides as rapidly as possible on the basis of results from numerous optimization experiments, taking both price and simplicity of the method into consideration. Results of these optimization experiments are summarized in the following text. The term "solid phase oligonucleotide" is used in this text instead of "solid phase primer" since the solid phase bound oligonucleotide does not function as a primer unless the DIAPOPS procedure is carried out.
The measurement of the amount of solid phase bound oligonucleotide is performed using hybridization detection ( ![]() ). This measurement only yields a relative amount of solid phase bound oligonucleotide. In order to obtain a truly quantitative result, radioactively-labelled oligonucleotides should be used (
). This measurement only yields a relative amount of solid phase bound oligonucleotide. In order to obtain a truly quantitative result, radioactively-labelled oligonucleotides should be used ( ![]() ). However, for optimization purposes, the hybridization detection is adequate, and all experiments for optimization of the covalent binding procedure have been carried out using this approach.
). However, for optimization purposes, the hybridization detection is adequate, and all experiments for optimization of the covalent binding procedure have been carried out using this approach.
2. Time of incubation during the covalent binding
The standard time of incubation for the covalent binding is 5 hours at 50ºC. This period of time has been selected on the basis of several criteria. As seen on the time curve illustrated in Figure 1, a plateau is reached after 2 hours, and after this no more solid phase oligonucleotides are bound to the surface of the NucleoLink Strips. The plateau remains constant for 24 hours. However, this experiment has been carried out repeatedly, and most results are not as clear cut as shown in Figure 1. Figures 2-4 illustrate that the amount of solid phase bound oligonucleotides can increase after incubation for 2 ½ hours (150 minutes) up 5 hours (300 minutes). In particular, the results in Figure 4 shows two identical experiments carried out using the standard DIAPOPS binding procedure ( ![]() ). These curves indicate that the highest amount of solid phase oligonucleotides was not attained after 5 hours. However, numerous results have indicated that there is no increase in the amount of solid phase bound oligonucleotides after incubation of 5 to 24 hours and that the plateau is probably reached after 4 hours. As indicated in the DIAPOPS procedure (
). These curves indicate that the highest amount of solid phase oligonucleotides was not attained after 5 hours. However, numerous results have indicated that there is no increase in the amount of solid phase bound oligonucleotides after incubation of 5 to 24 hours and that the plateau is probably reached after 4 hours. As indicated in the DIAPOPS procedure ( ![]() ), the incubation time can be chosen for convenience to be from 4 to 24 hours.
), the incubation time can be chosen for convenience to be from 4 to 24 hours.
3. Temperature of incubation during the covalent binding
The effect of the incubation temperature during covalent binding on the amount of solid phase bound oligonucleotides was measured as a function of the incubation time. The results are shown in Figure 2. The amount of solid phase bound oligonucleotide increases as a function of the incubation temperature and, as seen in Figure 1, also as a function of the incubation time. There is no significant difference in the amount of solid phase bound oligonucleotide with the two highest temperatures tested, 55ºC and 60ºC. This indicates that the temperature yielding the highest amount of solid phase bound oligonucleotides has been reached with these temperatures. Furthermore, there is only little difference in the amount bound in the experiments where temperatures between 45ºC and 60ºC are used.
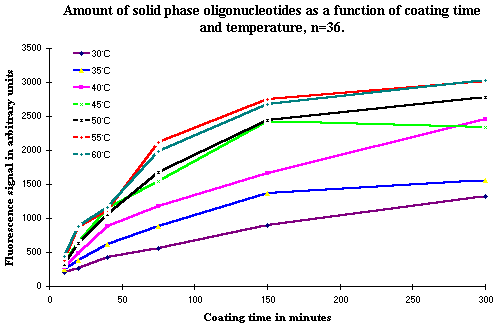 Figure 2: Shows the amount of solid phase bound oligonucleotides measured by hybridization as a function of incubation time and temperature. The increase in temperature closely follows the increase in the amount of solid phase oligonucleotide as a function of the incubation time. |
3 a). Selection of 50ºC
The selected temperature of 50ºC is a compromise. This temperature does not yield the highest amount of covalently bound solid phase oligonucleotides, but there is only a very limited difference between this temperature and the higher temperature of 55ºC. The reason for the choice of 50ºC is that other steps in the DIAPOPS procedure also require 50ºC. It simplifies the procedure greatly when the need for different incubation temperatures is reduced. In the DIAPOPS procedure only room temperature and 50ºC are necessary. If a higher temperature than 55ºC or 60ºC is available, these higher temperature can be used, and a shorter incubation time can be used. However, the incubation time should be tested and optimized in systems using a temperature different from 50ºC.
4. Incubation during the covalent binding with or without shaking
The incubation time could be reduced if the rate of the covalent binding of the phosphorylated oligonucleotide increased when the incubation was performed with shaking. To determine if this were the case, an experiment was conducted comparing the amount of solid phase bound oligonucleotide as a function of incubation time with and without shaking. Results from this experiment are shown in Figure 3. There is no indication of any difference in the results between the two experimental procedures, and therefore, one may conclude that the rate of covalent binding cannot be increased by using incubation with shaking.
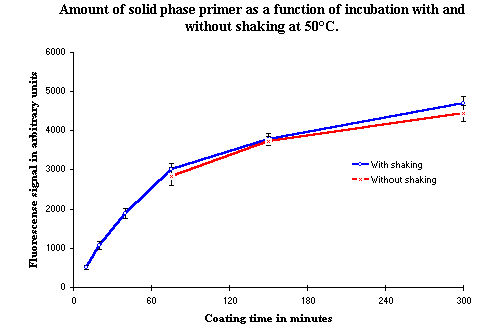 Figure 3: The amount of solid phase bound oligonucleotides as a function of time, with and without shaking. The first three points are missing from the curve without shaking due to experimental problems. However, this curve follows very closely the curve of the results from the experiment incubated with shaking. |
5. EDC concentration in the covalent binding
The standard concentration of EDC used in the DIAPOPS procedure is 10 mM ( ![]() ). This concentration has been selected on the basis of results of which some are shown in Figure 4. From these results it is obvious that the concentration of EDC can be decreased to 5 mM or be increased to 12.5 mM without causing any significant change in the amount of solid phase bound oligonucleotide. At 100 mM, the amount is very low. Furthermore, when the concentration is increased to 16 or 20 mM, the amount of solid phase bound oligonucleotide drastically decreases. This is illustrated in Figure 5, where additional 25 µl of EDC with a concentration of 40 mM will increase the concentration to 16 mM, and 50 µl will increase the concentration to 20 mM.
). This concentration has been selected on the basis of results of which some are shown in Figure 4. From these results it is obvious that the concentration of EDC can be decreased to 5 mM or be increased to 12.5 mM without causing any significant change in the amount of solid phase bound oligonucleotide. At 100 mM, the amount is very low. Furthermore, when the concentration is increased to 16 or 20 mM, the amount of solid phase bound oligonucleotide drastically decreases. This is illustrated in Figure 5, where additional 25 µl of EDC with a concentration of 40 mM will increase the concentration to 16 mM, and 50 µl will increase the concentration to 20 mM.
5 a). The importance of controlling the EDC concentration
In Figure 5, binding experiments with EDC concentrations of 16 and 20 mM are presented. These two concentrations are important, because in the previously used method for covalently binding the phosphorylated oligonucleotide to the surface of NucleoLink Strips, ( ![]() ), the oligonucleotides and the EDC were added separately. The oligonucleotides were added to each well in the NucleoLink Strips in 75 µl buffer as the first step in the binding procedure. Following this, EDC was added in 25 µl buffer with a concentration of 40 mM. This method has the advantage of not allowing the EDC to react with the oligonucleotides outside the NucleoLink wells. Unfortunately, it also has the disadvantage of introducing the possibility of adding EDC twice, as it is difficult to see whether EDC has been added or not. If the operator is interrupted during the addition of the EDC, it is almost impossible to distinguish between the wells with 75 µl of clear liquid without EDC and the wells with 100 µl already containing EDC. In this case, most people will choose to be sure and add an extra amount of 25 µl EDC to the questionable wells. As seen in Figure 5, this will drastically decrease the amount of solid phase bound oligonucleotides. This decrease in solid phase bound oligonucleotides will significantly reduce DIAPOPS results.
), the oligonucleotides and the EDC were added separately. The oligonucleotides were added to each well in the NucleoLink Strips in 75 µl buffer as the first step in the binding procedure. Following this, EDC was added in 25 µl buffer with a concentration of 40 mM. This method has the advantage of not allowing the EDC to react with the oligonucleotides outside the NucleoLink wells. Unfortunately, it also has the disadvantage of introducing the possibility of adding EDC twice, as it is difficult to see whether EDC has been added or not. If the operator is interrupted during the addition of the EDC, it is almost impossible to distinguish between the wells with 75 µl of clear liquid without EDC and the wells with 100 µl already containing EDC. In this case, most people will choose to be sure and add an extra amount of 25 µl EDC to the questionable wells. As seen in Figure 5, this will drastically decrease the amount of solid phase bound oligonucleotides. This decrease in solid phase bound oligonucleotides will significantly reduce DIAPOPS results.
5 b). Extra addition of EDC during incubation
EDC is a very reactive compound ( ![]() ), and although the use of 1-methylimidazole as the incubation buffer stabilizes the reaction (
), and although the use of 1-methylimidazole as the incubation buffer stabilizes the reaction ( ![]() ), there is a possibility that EDC can be the limiting factor in the binding reaction and therefore may cause a decrease in the rate of the covalent binding of the phosphorylated oligonucleotide. For this reason, the effect on the rate covalent binding of adding fresh EDC during incubation was examined. Results from this experiment are presented in Figure 6. From the results it is obvious that additional EDC does not improve the binding reaction, but can cause a significant decrease in the rate of binding.
), there is a possibility that EDC can be the limiting factor in the binding reaction and therefore may cause a decrease in the rate of the covalent binding of the phosphorylated oligonucleotide. For this reason, the effect on the rate covalent binding of adding fresh EDC during incubation was examined. Results from this experiment are presented in Figure 6. From the results it is obvious that additional EDC does not improve the binding reaction, but can cause a significant decrease in the rate of binding.
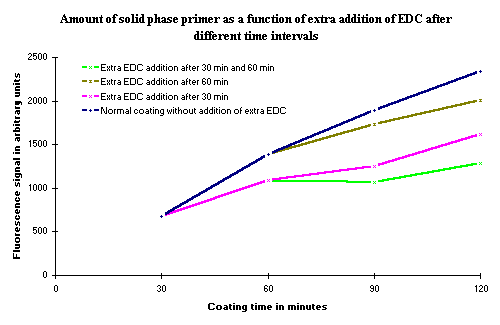 Figure 6: Amount of covalent bound solid phase oligonucleotide as a function of the coating time. The different curves represent the amount of solid phase oligonucleotide after addition of 25 µl 40 mM EDC at the times indicated. As seen, the only effect of an extra addition of EDC is a decrease in the amount of covalently bound oligonucleotide. |
6. Concentration of the phosphorylated oligonucleotide
The concentration of the phosphorylated oligonucleotide which is to be covalently bound to the surface of the NucleoLink Strips is experimentally selected to be 100 ng added to each well in the NucleoLink Strips. This yields about 20 ng bound to each well ( ![]() ).
).
The concentration of 100 ng per well was selected from experiments using the hybridization method to examine whether a higher concentration of phosphorylated oligonucleotides would cause more oligonucleotides to bind to the surface of the NucleoLink Strips. Results from the experiment presented in Figure 7 indicate that the rate of covalent binding is increased with the higher oligonucleotide concentration of 1000 ng per well. However, the total amount bound after 5 hours of incubation is not altered.
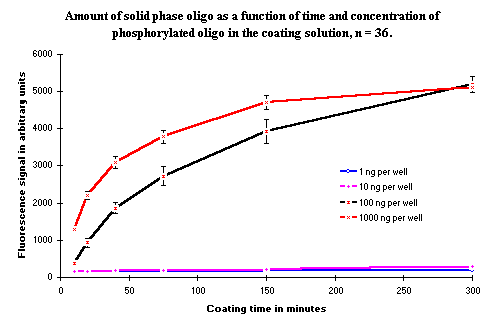 Figure 7: Amount of covalent bound solid phase oligonucleotide as a function of time. Four different concentrations of the phosphorylated oligonucleotide have been analyzed. The two lowest concentrations, 1 and 10 ng/well, do not give any signal in this experiment. A higher probe concentration was also tested to ascertain whether any solid phase oligonucleotides could be detected at these concentration. The results showed that with both 1 and 10 ng probe, the solid phase oligonucleotides could be detected with the higher concentration of biotinylated probe. |
When 1 ng and 10 ng phosphorylated oligonucleotide were added per well of, no signal was observed with the used concentration of hybridization detection probe. This is because the selected concentration of biotinylated probe is too low to detect the low amount of oligonucleotide bound to the surface ( ![]() ). In a second hybridization, the concentration of biotinylated probe was increased. The results (results not shown) from the hybridization with a higher concentration of the biotinylated probe indicated that the binding rate of oligonucleotides with concentrations of 1 ng and 10 ng were comparable to the concentration observed in Figure 7, but since a higher probe concentration was necessary to detect the solid phase bound oligonucleotides, the total amount of bound oligonucleotide is lower.
). In a second hybridization, the concentration of biotinylated probe was increased. The results (results not shown) from the hybridization with a higher concentration of the biotinylated probe indicated that the binding rate of oligonucleotides with concentrations of 1 ng and 10 ng were comparable to the concentration observed in Figure 7, but since a higher probe concentration was necessary to detect the solid phase bound oligonucleotides, the total amount of bound oligonucleotide is lower.
7. Incubation Buffer and pH of buffer
The buffer used in the DIAPOPS procedure for covalent binding the phosphorylated oligonucleotides to the surface of the NucleoLink Strips is 1-methylimidazole ( ![]() ) at pH 7.0. Different pH-settings of the 1-methylimidazole buffer were tested. Figure 8 illustrates the results of these experiments. It is obvious that when pH increases, the amount of solid phase bound oligonucleotide is not affected to any great extend. When the pH is lowered there is a significant decrease in the amount of covalently bound oligonucleotide. Even the use of unadjusted 1-methylimidazole buffer, which has a high pH, does not affect the result to a great extend. An error in this step, i.e. omitting to adjust the pH, will have little effect on the results.
) at pH 7.0. Different pH-settings of the 1-methylimidazole buffer were tested. Figure 8 illustrates the results of these experiments. It is obvious that when pH increases, the amount of solid phase bound oligonucleotide is not affected to any great extend. When the pH is lowered there is a significant decrease in the amount of covalently bound oligonucleotide. Even the use of unadjusted 1-methylimidazole buffer, which has a high pH, does not affect the result to a great extend. An error in this step, i.e. omitting to adjust the pH, will have little effect on the results.
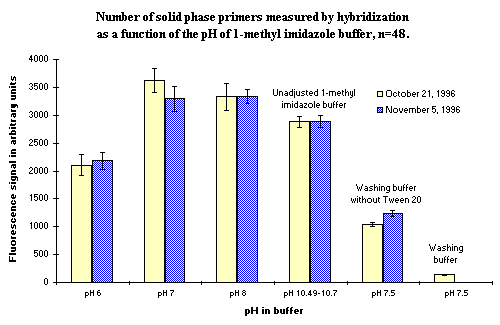 Figure 8: Amount of covalent bound solid phase oligonucleotide as a function of different buffers. The unadjusted 1-methylimidazole buffer and the normal washing buffer are also included in the experiment. It is clear that failing to adjust the pH of the 1-methylimidazole buffer does not affect the results much, but the use of a wrong buffer may significantly decrease the amount of solid phase bound oligonucleotides. |
The use of the normal washing buffer as coating buffer was also tested in the same experiment. This was in order to examine the consequence of an error in this step if the most frequently used buffer in the DIAPOPS procedure, the washing buffer, was used as the coupling buffer. If the washing buffer contains Tween 20, no oligonucleotide will be bound to the surface of the NucleoLink Strips. If the buffer is used in the absence of Tween 20, there is a binding of aproximately 30% of the normal level.
8. Conclusion
The conditions in the DIAPOPS procedure for covalent binding of a phosphorylated oligonucleotide to the surface of NucleoLink ( ![]() ) were selected to bind the highest amount of oligonucleotide as rapidly as possible, and taking both price and simplicity of the method into consideration. The time of incubation can be decreased by increasing the incubation temperature or increasing the concentration of the phosphorylated oligonucleotide. However, the amount of solid phase bound oligonucleotide will not be increased. The concentration of EDC must be carefully controlled, because both too high and too low concentrations will decrease the amount of solid phase oligonucleotide. A high pH in the incubation buffer will not affect the binding, but a lower pH has an inhibitory effect.
) were selected to bind the highest amount of oligonucleotide as rapidly as possible, and taking both price and simplicity of the method into consideration. The time of incubation can be decreased by increasing the incubation temperature or increasing the concentration of the phosphorylated oligonucleotide. However, the amount of solid phase bound oligonucleotide will not be increased. The concentration of EDC must be carefully controlled, because both too high and too low concentrations will decrease the amount of solid phase oligonucleotide. A high pH in the incubation buffer will not affect the binding, but a lower pH has an inhibitory effect.