TechNote Volume 3 No. 16 - Quality Control of NucleoLink™ Strips
Revised Version May, 96
Thin walled NucleoLink Strips (Cat. No. 248259) facilitate solid phase PCR*. The Strips are made of an activated heat stable polymer and support covalent binding of DNA by carbodiimide mediated condensation. One type of solid phase PCR, the process of extending immobilized primers during amplification, is the DIAPOPS assay ![]() )
)![]() )
)![]() )
)
The DIAPOPS assay is simplified as "ELISA like" procedures (e.g. labelled probes, substrates, and instruments can be used). Since different functional groups (biotin, digoxigenin) or enzymes can be coupled to DNA, no limitation in "ELISA like" procedures is caused by utilizing DNA. NucleoLink Strips can be assembled in MicroWelI™ format ![]() )
)
Since the strips are compatible with the DIAPOPS procedure, this technique is used to test the performance of the strips.
The DIAPOPS Technique

Methods ![]() )
)
Binding of Primer ![]() )
)
100 ng/well of 5' phosphorylated downstream primer is dissolved in 10 mM 1 -methyl-imidazole (1 -Melm), pH 7.0, and 75 pi is pipetted into each well. Then 25 µl freshly made 40 mM 1-ethyl-3-(3-dimethyiaminopropyl) carbodiimide dissolved in 10 mM 1-MeIm is added to each well.The NucleoLink™ Strips are sealed with Tape 8 and are incubated at 50ºC for 5 hours.
After incubation, the NucleoLink Strips are washed with 0.4 N NAOH, 0.25% Tween 20 at 50ºC ![]() )
)
Amplification ![]() )
)
Although the DIAPOPS procedure greatly diminishes the risk of contaminating the laboratory with amplicon DNA, all PCR reactions are prepared in a laboratory separated from the QC laboratory. After preparation, the reactions are transported to the QC laboratory for amplification and detection.
To block the wells before amplification, add to each well 200 µl of 100nM TRIS-HCl (pH 7.5), 150nM NaCl, and 0.1% Tween 20 with 10mg/ml BSA ![]() )
)![]() )
)![]() )
)
5 µl template DNA (10-22 - 10-16 moles/µl) is mixed with 45 µl PCR solution consisting of buffer (10 mM Tris-HCl (pH 8.3),50 mM KC], 0.1 % Tween 20), 1.8 mM MgCl2, 200 µM of each DNTP, 0.5 µM upstream primer, 0.06 µM downstream primer, and 1 U Taq-polymerase.
Thermocycling
The wells are sealed with Tape 8 in order to avoid evaporation during amplification ![]() )
)![]() )
)
Since amplification is performed in a thermal cycler with a heated lid ![]() )
)
NucleoLink™ Strips are washed three times, incubated for five minutes at room temperature and washed three times more with freshly made 0.2 N NaOH, 0.1 % Tween 20 ![]() )
)
Detection ![]() )
)
Hybridization is performed for one hour with 50 nM biotinylated detection probe at 50ºC in 100 µl hybridization buffer (5 x SSC, 0.1 % Tween 20, 0.5% Blocking Reagent). After hybridization, the strips are washed three times, incubated for 15 minutes at 50ºC and washed three times more with 0.5 x SSC, 0.1 % Tween 20.
Streptavidin-alkaline phosphatase conjugate ![]() )
)
Finally, the strips are incubated for 30 minutes at 50ºC with 100 µl 1 mM 4-methylumbelliferyl phosphate ![]() )
)![]() )
)
Certification
The results are plotted with the initial template concentration on the x-axis and the fluorescence intensifies obtained on the y-axis (see figure 2 below).
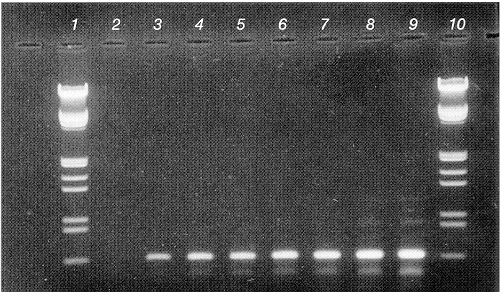 Figure 1: Solution phase PCR on cloned bovine leukemia virus (BLV |
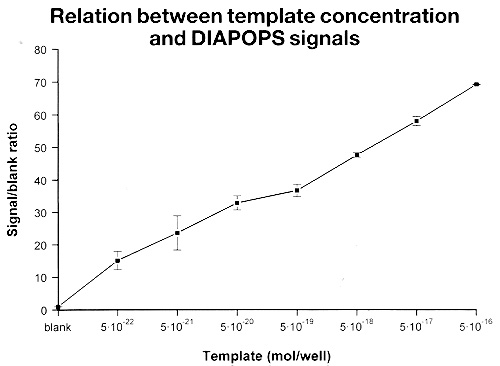 Figure 2: DIAPOPS detection corresponding to the gel shown in figure 1 using the procedure described in the text. In order to be certified the NucleoLink™ Strips should support results where the relation ship between initial template concentration and obtained fluorescence intensity is linear with a correlation coefficient of 0.9 or better, for at least 4 orders of magnitude. |
Reference:
Rasmussen, Soren R. et al, »Combined Polymerase Chain Reaction Hybridization Microplate Assay Used to Detect Bovine Leukemia Virus and Salmonella«, Clin. Chem. 40/2, 200-205 (1994).