A successful DIAPOPS analysis depends on a successful solid phase DNA amplification
1. Introduction
A successful DIAPOPS analysis depends on a successful solid phase DNA amplification ![]() )
)![]() )
)![]() )
)![]() )
)![]() )
)![]() )
)
2. Methods for measuring the amount of solid phase bound oligonucleotides
The measurement of the solid phase bound oligonucleotides on the surface of NucleoLink Strips can be carried out using several different methods. In the following text the term "solid phase oligonucleotide" is used instead of "solid phase primer" since it does not function as a primer unless the DIAPOPS procedure is carried out. Three different methods have been tested at the Nunc A/S Research Laboratory.
3. Radioactively-labelled oligonucleotides
An absolute quantitative measurement of the amount of solid phase bound oligonucleotide can be made when a radioactively-labelled oligonucleotide is used, whereas the methods using a biotinylated solid phase oligonucleotide only yields a relative result which can be compared to a standard.
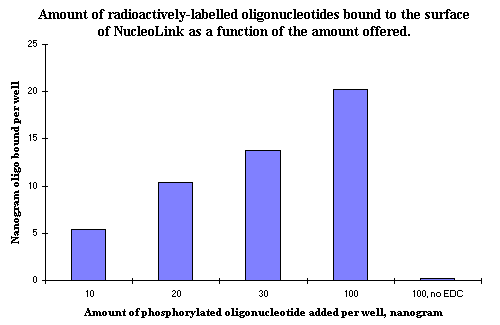 Figure 1: The amount of oligonucleotide covalently bound to the surface of NucleoLink after EDC mediated binding. The different amounts offered to each well show that a decreasing percentage is bound, but that an increasing total amount is bound to the surface. Addition of 100 ng of phosphorylated oligonucleotide results in approximately 2 pmol oligonucleotide being covalently bound to the surface of one NucleoLink well. |
A measurement of nanogram amounts of oligonucleotides covalently bound to the surface of NucleoLink Strips is shown in Figure 1. The quantification is made by adding a small unknown amount of radioactively-labelled oligonucleotide to a much larger, known amount of non-radioactive oligonucleotide.
The concentration of the radioactively labelled oligonucleotide is very low. Therefore, there is no change in the total concentration of oligonucleotide when the labelled oligonucleotide is added to the 100 ng unlabelled oligonucleotide offered to each well. For this reason, the proportion of radioactive counts bound to the NucleoLink Strips also yields the proportion of the total amount of non-radioactive oligonucleotides bound. When 100 ng is added to each well, the resulting amount of covalently bound oligonucleotide is approximately 20 ng. This is approximately 2.5 pmol per well of an oligonucleotide nucleotide of 25 bases.
4. Biotin-labelled solid phase oligonucleotide
In this procedure a 5'-phosphorylated oligonucleotide was also 3'-biotinylated. The oligonucleotides covalently bound to the NucleoLink surface could be directly detected using streptavidin conjugated with alkaline phosphatase. This method can yield the same optimization results as the radioactive method, but the results can not be used to produce an absolute quantification of the amount of oligonucleotides bound. The method can be used, but it does not give any indication of the usability of the DNA, because the oligonucleotides might be bound in such a way as to inhibit the hybridization which is the initial step in the PCR ![]() )
)
5. Hybridization with a labelled probe to the solid phase bound oligonucleotide
Annealing to the solid phase oligonucleotide acting as a primer is the initial step of the solid phase DNA amplification ![]() )
)
5 a). A mix of hybridization probes
A normal hybridization procedure was used for the analysis, but the hybridization solution contained both biotinylated and unbiotinylated hybridization probes. This was done to ensure that the surface was saturated with probes. The concentration of the biotinylated oligonucleotide has to be low (approximately 50 pM 5 fmol per well) in order to obtain a signal low enough to be dynamic. If the biotinylated oligonucleotide is used alone at this low concentration, a change in the number of solid phase oligonucleotides would not automatically give a change in signal. The reason for this is that if 2.5 pmol solid phase oligonucleotide is bound to the surface, there will be more solid phase oligonucleotides than probes. A shift in the number of solid phase bound oligonucleotides would not automatically bring about a change in the amount of captured oligonucleotides, and thus there would be no change in signal.
This problem is solved when an unlabelled probe is used together with the labelled probe. A total concentration can be chosen which is high enough to saturate the surface, and the concentration of the biotinylated oligonucleotide will still be low enough to give a dynamic signal. The concentration of the biotinylated probe was experimentally selected to yield a signal of 5000 arbitrary fluorescence units, as shown in Figure 2.
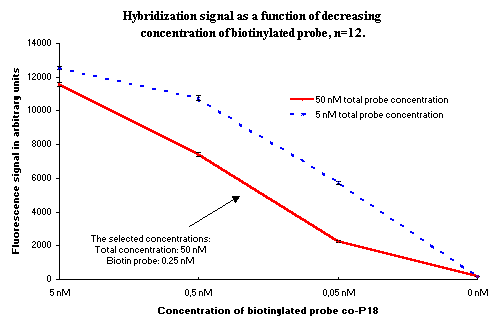 Figure 2: Fluorescence signal as a function of concentration of biotin-labelled probe in two different concentrations of labelled and unlabelled probe. The arrow shows the concentration selected for use in the experiments for studying the binding of DNA to the NucleoLink surface. |
5 b). Preheating of the coated NucleoLink Strips
Before analysis of the solid phase bound oligonucleotide by hybridization the coated wells were heated to 94ºC for 20 minutes with 100 µl DIAPOPS wash buffer ![]() )
)![]() )
)
6. Correlation between the number of solid phase primers and the DIAPOPS signal
To confirm the hypothesis that the number of oligonucleotides is directly related to the solid phase DNA amplification and therefore to the DIAPOPS signal, a comparison of the amount of oligonucleotides bound to the solid phase and the solid phase DNA amplification was carried out. The amount of solid phase oligonucleotides was adjusted by controlling the time of covalent binding ![]() )
)![]() )
)![]() )
)![]() )
)
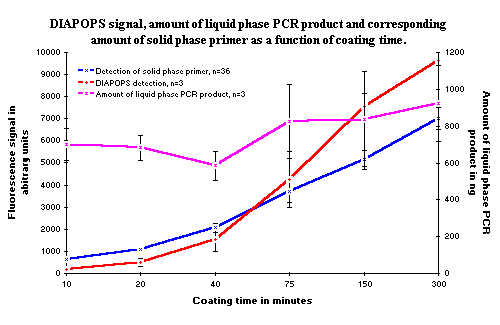 Figure 3: The solid phase DNA amplification (red line) as a function of the amount of solid phase oligonucleotides. The amount of solid phase oligonucleotides (blue line) was adjusted by controlling the time of the covalent binding. The amount of liquid phase PCR product (purple line) in the reactions is approximately constant. The increase in the amount of liquid phase PCR product is accidental, and is not correlated to the rising amount of solid phase oligonucleotides. All data from the solid phase detection using hybridization have been multiplied by two, in order to be comparable with the DIAPOPS data. |
7. Conclusion
The solid phase DNA amplification is very dependent on the number of solid phase primers. The relative amount of solid phase oligonucleotides which can act as primers in the solid phase amplification can be estimated using detection by hybridization.